TGsol (Translational Genomics for Solanaceae)
TGsol (Translational Genomics for Solanaceae) is the specialized web interface providing the integrated view of genomic big-data available to practical breeding process of Solanaceae crop. TGsol will play important role as a bridge between genomic evidence and molecular breeding via supporting to effective communication and utilization.
VISION (Aims)
· Sharing the Genomic big-data
· Integrating front (genome) industry and rear (breeding) industry (market)
· Invigorating BREEDING · SEED industry
TGsol Function (Summary)
·
Target Crop: Solanaceae crop (Tomato, Pepper and Potato)
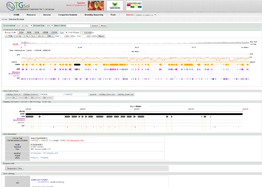
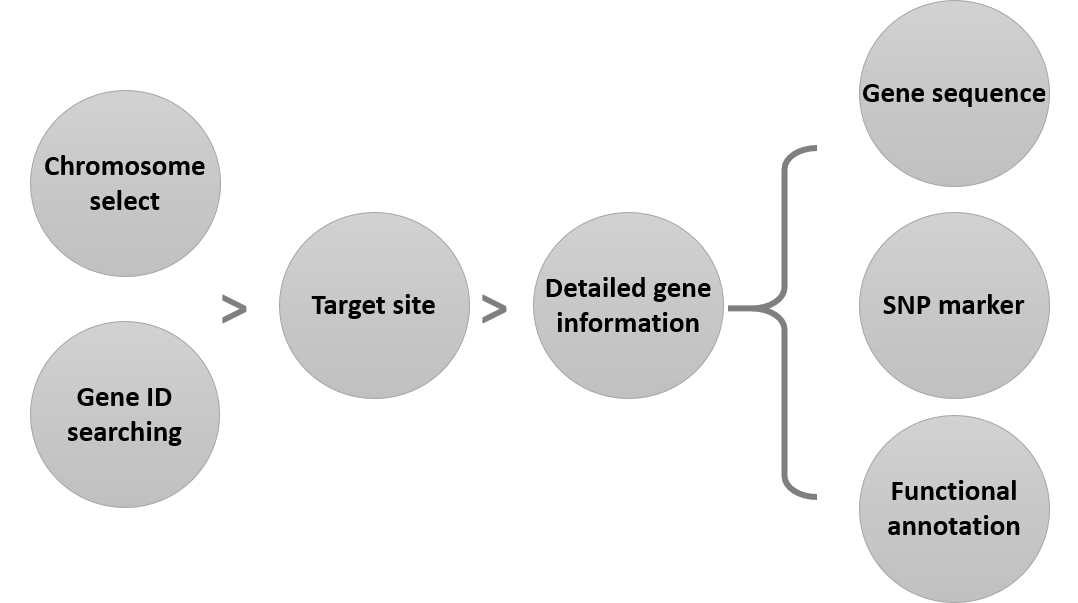
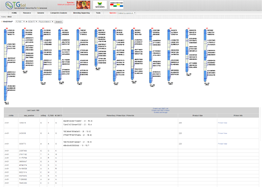
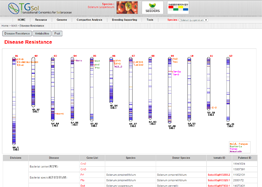
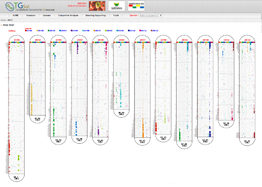
Main Function : Genome browser, Breeding supporting viewer (MAB, MAS), Comparative analysis tool (viewer), BLAST
Application : TGsol provide re-processed and re-interpreted information desired for marker-assisted breeding process.
 TGsol
TGsol
 Solanum lycopersicum
Solanum lycopersicum
 Solanum tuberosum L.
Solanum tuberosum L.
 Capsicum annuum L.
Capsicum annuum L.
 Comparative Analysis
Comparative Analysis
 Forums
Forums 







 Contact Us : help@seeders.co.kr +82 042-710-4025
Contact Us : help@seeders.co.kr +82 042-710-4025